Molecule and surface display style can be changed on a per-molecule, per-surface basis, it is possible to change atom and bond display style, color and size. It is also possible to change the display style for residues and show/hide the dipole moment.
To quickly change the molecular display style use the Style toolbar menu button.
All the per-molecule display settings can be changed through the Molecule Display Settings dialog (Display->Molecule).
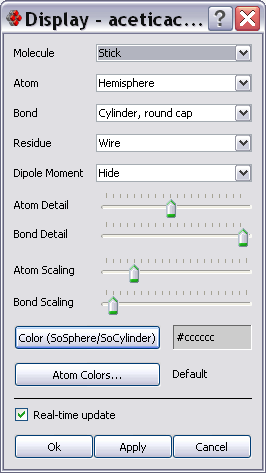
The overall molecule representation can be changed through the Molecule drop down listbox; available styles are:
- CPK (Spacefill)
- Ball and Stick
- Ball and Wire
- Stick
- Wireframe
Atom representation can be changed through the Atom drop-down listbox; some of the available representations are:
- Hemisphere - use a hemisphere facing the point of view
- gluSphere - use OpenGL gluSphere (use this when exporting to PostScript/PDF)
- SoSphere - use OpenInventor SoSphere: draws all the atoms with the same color selected with the Color button on the same dialog box.
- Billboard - use point sprites to render atoms, useful with big molecules; use this style with CPK molecule rendering style.
- Level of Detail - complexity of the rendered geometry changes depending on the distance from the point of view
Bond representation can be changed through the Bond drop-down listbox; some of the available representations are:
- Cylinder
- Cylinder, round cap - use this in conjunction with Stick molecule rendering syle to display the molecule in Liquorice mode.
- gluCylinder - use OpenGL gluCylinder (use this when exporting to PostScript/PDF)
- SoCylinder - use OpenInventor SoCylinder: draws all the bonds with the same color selected with the Color button on the same dialog box.
- Level of Detail - complexity of the rendered geometry changes depending on the distance from the point of view
Residue representation can be changed through the Residue drop-down listbox; available styles are:
- Wire
- Stick
- Line Ribbon
- Flat Ribbon
- Solid Ribbon
- Schematic
Atom and bond detail can be changed with the Detail sliders.
Size of atoms and bonds can be changed with the Scaling sliders. The minimum value for atom and bond radius is zero; the maximum value of atom radius is the length of the covalent radius.
The color of atom and bonds rendered with SoSphere and SoCylinder style can be selected by pressing the Color button. When a SoXXX style is selected all the atom and/or bonds are rendered with the same color.
Hiding atoms and bonds
To display only surfaces or other features (e.g. protein ribbon) without atoms and bonds simply set the atom and bond scaling factor to zero.
Changing the atom colors
Atom colors can be changed by forcing Molekel to read the per-atom color values from a text file, a sample file is included in the data directory.
The file format is a list of red green blue values between 0 and 1 with the line number matching the atomic number.
E.g.
0.00 0.00 1.00 0.00 1.00 0.00 0.70 0.30 0.00 0.40 1.00 0.33 ...
In this case the first line is the color of H atoms the second line the color of He atoms and so on.
Colors must be applied on per-molecule basis and are not persistent i.e. the next time you load the same molecule you have to re-load the atom color file.
Applying changes
When the Real-time update checkbox is checked changes are applied immediately and the 3d view refreshed each time a value is changed.