Molekel can export animation and animate all the visible molecular surfaces by recomputing the surfaces at each animation step. Animation can be exported as individual frames or AVI files (on Windows).
When exporting animations to individual frames the user needs to select the directory where the individual frames will be written. Files are exported as png images with names which include the frame sequence number.
To create animations from the generated frames you can use a program like
Sample mencoder command line which will generate an mpeg video m.mpeg from the png images found in c:\tmp\molekelvideo1 :
mencoder.exe mf://c:\tmp\molekelvideo1\*.png -o m.mpg -ovc lavc -lavcopts vcodec=mpeg2video -of mpeg
The frame rate should match the frame rate specified in Molekel's Export Animation dialog.
To specify the frame rate on mencoder command line use the -ofps switch.
To export an animation open the Export Animation dialog by selecting the Animation-Export Animation menu item, modify the parameter values, click on Ok and select the output directory (individual frames) or file (AVI).
Export Animation Dialog
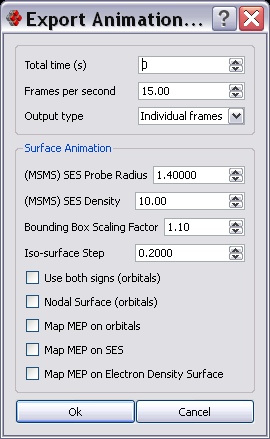
This dialog box allows the user to speficy duration, frames per second and output type (individual frames or AVI) and the parameters that will be used for rendering the visible surfaces while animating.
Note that:
- the values specified here will override any previously specified value and will apply to all the visible surfaces
- the same step will be used for all the visible iso-surfaces
- the same value of 0.05 will be used for all the visible iso-surfaces (i.e. the suface scalar_field( x, y, z ) = 0.05 will be computed)
The labels on this dialog should give a good explanation of what the parameters other than Bounding Box Scaling Factor are.
Bounding Box Scaling Factor is the value by which the size of each molecule's bounding box is multiplied before computing the isosurfaces (if any).
Having only one set of values that apply to all the surfaces is definitely not an optimal solution; the only way to cover all the possible options (different bounding boxes, animating a surface by changing an iso-value or probe radius at each frame etc.) is to introduce scripting, which will be added in the next version (5.1) of Molekel.